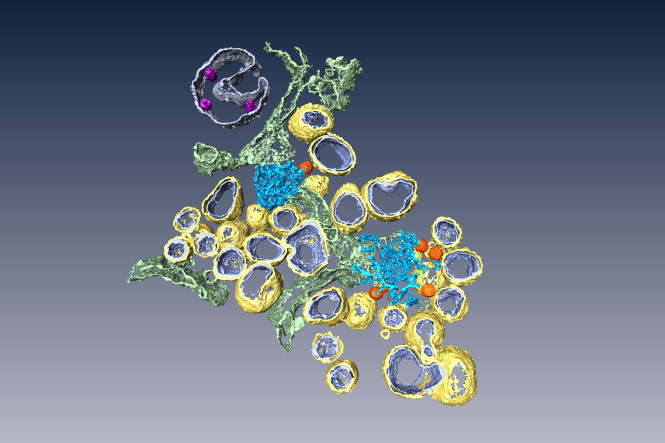
One of the most fascinating aspects in the life cycle of coronaviruses is the dramatic remodeling of intracellular membranes that takes place in infected cells. Along the years, an assortment of coronavirus-induced membrane structures, with intriguing variations per coronavirus group, have been described in the literature. These membrane structures were thought to support viral genome replication and have been collectively referred to as the coronavirus replication organelle. However, despite much speculation, it was unclear which of these structures were actively involved in viral RNA synthesis.
This work, carried out in collaboration with the group of Prof. Eric Snijder in the Medical Microbiology Department, settles this long-debated issue and provides a unifying model of the coronavirus replication organelle. Combining 3D and 2D EM with new sample preparation protocols, we demonstrated that coronaviruses across different genera induce essentially the same type of membrane structures. Using EM autoradiography, we were able to track down the site of viral RNA synthesis to the virus-induced double-membrane vesicles (DMVs). Our results, published in PLoS Biology and highlighted in Nature Reviews Microbiology, clearly establish DMVs as the central hub for viral RNA synthesis and, therefore, as a potential drug target in coronavirus infection.
Eric J Snijder, Ronald WAL Limpens, Adriaan H de Wilde, Anja WM de Jong, Jessika C Zevenhoven-Dobbe, Helena J Maier, Frank FGA Faas, Abraham J Koster, Montserrat Bárcena. “A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis” PLoS Biol. 2020 Jun 8;18(6):e3000715
https://doi.org/10.1371/journal.pbio.3000715
Highlight in Nature Reviews Microbiology (June 19):