Over time the work on MHC class I antigen presentation shifted more to MHC class II antigen presentation and the cell biology of the endosomal system. There are many questions that are studied in the Neefjes lab. They involve the control of maintenance and transport of late endosomes and bacteria containing phagosomes, the dynamics within a multivesicular body, the role of DUBs and kinases in the control of the endosomal system and the involvement of different motor proteins in the complex bidirectional movement of (MHC class II containing) late endosomes. Within the department of Cell and Chemical Biology (CCB), we have a host of high-tech techniques that support our research. These include mass spec of the ubiquitinome, chemistry to produce inhibitors and fluorophores, high-end electron and light microscopy and virology. In addition, the CCB harbors the Leiden chemical screening and protein facility that allow top notch biochemistry.
From the work on the endosomal system, the other two themes developed and the technologies used are overlapping. These two themes have as a major goal the translation into clinical practice. Neefjes is directly involved in producing an anti-cancer drug at GMP grade and a clinical study that is in preparation. From the bacterial involvement in cancer, we expect that clinical guidelines can be developed to allow early diagnoses of patients at risk after such an infection. The Neefjes lab has a very broad and (to their own opinion) exciting portfolio of research.
The cell biology of the endosomal system and MHC class II antigen presentation
The Neefjes lab has identified various essential factors in control of late endosomal transport, fusion and retrofusion. They have shown how the dynein motor interacts with lysosomes and how this is controlled by the ER. In addition, they have identified another ER protein complex in control of the perinuclear location of the entire endosomal system. They are now expanding this work by placing various factors in the context of living cells in order to understand the dynamics of endosomal movement and fusion. CRISPR-based GFP and RFP knock-ins are made to follow the motor proteins on the moving vesicles. This work is further expanded to understand the biology and biochemistry of bidirectional movement towards the plasma membrane. This work is important for MHC class II antigen presentation, as well as a host of other processes such as autophagy, mitosis, host-cell infection and intracellular survival of bacterial pathogens. There are many other outstanding questions studied in the Neefjes lab. As the result of a multi-dimensional siRNA screen (Paul et al., Cell 2011), they identified various factors with interesting biology. These include the organizing principle of the endosomal system (Jongsma et al., Cell 2016) which yielded additional questions such as what are the associated proteins and how is the ultimate controller RNF26 retained in a perinuclear site in the ER. But it yielded also additional proteins that affected the endosomal system such as the DUB USP32 (with the Ovaa lab) and USP54 that controls EMT and transport processes. Other proteins that the lab is placing into the biology of the endosomal system include ER proteins in control of organelle contacts and proteins controlling motor protein recruitment. Within this cell biology, we are also interested in the dynamics within a multivesicular body and the leading question is whether the internal vesicles can fuse back to the limiting membrane. We have established a high-tech system to visualize this and are performing candidate approaches to decipher the molecular basis of ‘retrofusion’. This portfolio of endosomal research is sponsored by an ERC Adv grant and NWO grants.

Figure 1. The control of endosomal positioning by the ER ubiquitin ligase RNF26. All components have been identified in siRNA screens and protein pull-downs followed by mass spec activities at the LUMC, and confirmed and expanded by various cell biological and microscopical techniques. This new mechanism controls the intracellular positioning of the endosomal pathway and is a new concept in cell biology that also controls efficient timing of signal transduction, delivery of materials for lysosomal degradation, MHC class II antigen presentation and mitosis and probably many other processes involving the endosomal pathway.
The role of bacterial infections in cancer induction
Salmonella and other bacterial pathogens introduce factors in host cells to control infection and intracellular survival. We showed earlier (Kuijl et al. Nature 2007) that inhibition of the host kinase AKT1 was sufficient to eliminate intracellular bacteria, in fact defining the first antibiotic active through manipulation of host rather than pathogen proteins. Salmonella and other bacteria appeared to activate AKT1 during their infection cycle. Since AKT is also often activated in cancer, we wondered whether bacteria, through this mechanism, could contribute to cancer induction. Indeed, Salmonella infection was able to transform cells into cancer cells, when the cells already harbor one or two defined pretransforming mutations. This explains the link between chronic S.Typhi infections and the increased risk of gallbladder carcinoma, as observed in India. In fact, our lab results were confirmed in gallbladder carcinoma samples from India. These experiments (Scanu et al., Cell Host Microb 2015) also included a mouse experiment that showed that infection of APC+/- mice with another salmonella serovar, S.typhimurium, induced colon cancer. Since thise serovar is known from food poisoning, we wondered whether such food poisoning would increase the risk of colon cancer and started a nation-wide epidemiological study. We then showed that serious infections with –again- another salmonella serovar, S.enteritidis increases the risk of colon cancer almost 3-fold. We are studying these and other bacterial infections for their potential role in cancer induction by combining epidemiology, infection model systems and cell biology. This may yield clinical guidelines for patients with such infections to be enrolled in nation-wide cancer screening programs. This work is now a collaborative effort between the Dutch Health Institute (RIVM) and our lab, which providing unique epidemiological and clinical isolates for studying this connection. It is also supported by a shared grant from the Dutch Cancer Society KWF.
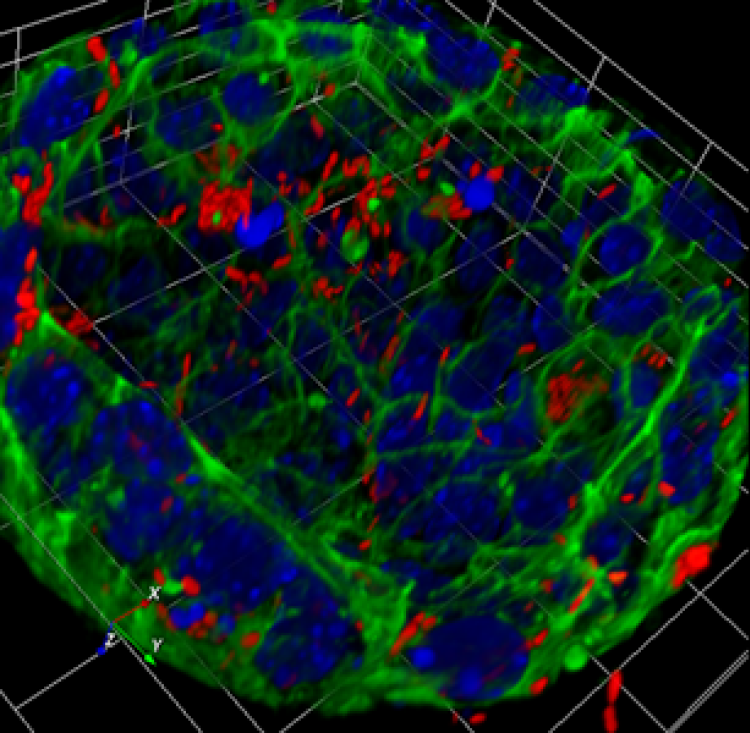
Figure 2. 3D-reconstruction of a gallbladder organoid transformed by infection with Salmonella typhimurium. Nuclei, blue; actin in green and Salmonella in Red
The cell biology of anthracycline anti-cancer drugs
From our cell biological studies, the Neefjes lab identified a new activity for the old anti-cancer drug doxorubicin, namely the induction of histone eviction from chromatin. This induces epigenetic programming and delays DNA repair (Pang et al., Nature Comm 2013), contributing to its anti-cancer activity. The lab continued by combining this and other variant drugs with epigenetic profiling, showing that doxorubicin and a variant drug called aclarubicin affect selective and different regions in the genome. Eviction of histones contributed to the anti-cancer activity. In fact, removal of DNA break formation still yielded powerful anti-cancer drugs but with strongly attenuated side effects. While doxorubicine induces cardiac failure, secondary tumors and infertility issues, some of these new variants were devoid of these (therapy limiting) side effects. Yet, these drugs have to be made at GMP level for further clinical testing and introduction in the clinic. One variant drug, aclarubicin is made at GMP level to start clinical trials in 2019 for relapsed AML patients and possibly a number of other cancer indications. The Neefjes lab will continue combining chemistry and cell biology to improve these drugs and to send these drugs to other areas of the genome. In addition, the lab is studying how the variant drugs act in chemo-immunotherapy options and affect immune responses in general. With a unique set of anthracycline variants, the Neefjes lab is in the best position to study the molecular details of drug interactions with DNA and chromatin using biophysical studies. Using modern technology to study the activity of old but well-working anti-cancer drugs visualizes new activities that can (sometimes at least) be chemically separated and explored for defined purposes.
Others
The Neefjes lab is also initiating new research to explore other fields related to cancer. They are studying proteostasis during mitosis with new ways of control for proteasome import. They are also studying the role of the host metabolome on the local microbiome in a multidisciplinary collaboration. And Neefjes has initiated an educational program aimed at preventing adolescent students from smoking.