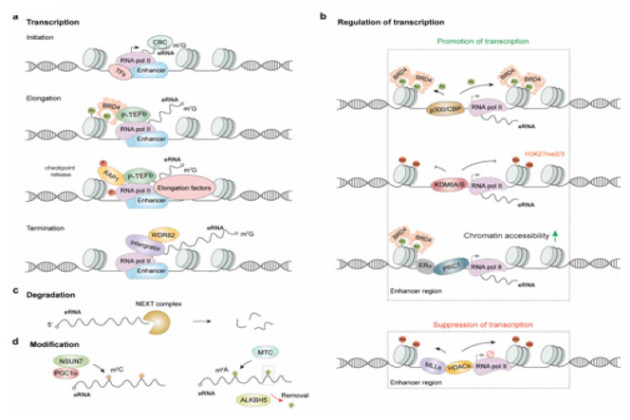
Ubiquitination is best known as a signal for proteasome-mediated protein degradation. Recent studies have uncovered new functions of ubiquitin and ubiquitin-like proteins. These functions signaling roles in DNA repair, immune responses and regulation of membrane dynamics.
Our lab aims to elucidate the function of ubiquitination and deubiquitination in these pathways. We have an interest in conjugating enzymes (E1-E2-E3’s) and deubiquitinating enzymes (DUBs) as therapeutic targets in diseases such as cancer and neurodegenerative diseases. Approaches include finding inhibitors of specific DUBs using state of the art high-throughput screening. It is our ultimate goal to validate specific targets with such small-molecule inhibitors of specific DUBs in cellular and animal models.
The van der Heden van Noort Lab

The ubiquitin (de)conjugation machinery is fiercely investigated the past two decades and for instance the basic dogma underlining the joint activity of E1, E2 and E3 enzymes to ligate ubiquitin to a target protein is widely accepted. Many of the intricacies of these systems however remain open for discussion and preparing new tools using a chemical approach to study ligase and hydrolase systems are essential for a better understanding. An example of understudied enzymatic activities is the cross-talk between ubiquitination and ADP-ribosylation. Bacterial effector proteins, such as the class of Legionella SidE enzymes, catalyze the covalent ADPribosylation of ubiquitin on Arg42, eventually leading to the phosphoribosyl ubiquitination of serine containing target proteins. As the matter-of-fact the interplay between ADPribosylation and ubiquitination is more prevalent in nature, in processes such as bacterial infection but also during DNA-damage responses in mammals, and we focus on studying such systems using a chemical biology approach.
The Mulder Lab
.jpg)
Ubiquitination most commonly leads to destruction of the modified protein, but can also change its activation, interactions or localization. These modifications are controlled by a complex enzymatic system, with more than 700 players in the ubiquitin network, and disruption within these systems could lead to diseases such as cancer and neurodegeneration. Detailed insights into the mechanism behind this is however yet to be obtained, thus slowing the development of targeted medicines for treatment of these diseases.
Much of our work aims to unravel the workings of these systems. Utilizing both activity-based probes as well as biochemistry and cell biology techniques, we are currently seeking to unravel the role of ubiquitination and the proteasome in Huntington’s disease, a dominantly inherited neurodegenerative disease. Moreover, we have a research interest in E3 ligases, the enzymes that attach ubiquitin to a substrate. Our research pursues the development of platform technologies to gain understanding on the biological and mechanistic information of E3 ligases, while capitalizing on insights to design strategies aimed at targeting specific E3 ligases. Together, this offers opportunities to exploit the ubiquitin system not only by small molecules but also by targeted protein degradation approaches as PROTACs. The success of the PROTAC strategy in targeting diverse pathological proteins relies, however, on the number and specificity of E3 ligases that can be recruited for this purpose. An exciting possibility to explore is the recruitment of E3 ligases with disease-specific expression or that are cell- or tissue- specific. This knowledge is exactly what is aimed for in my lab, along with the means (assays, reagents, probes) to reveal such knowledge.
The Geurink lab
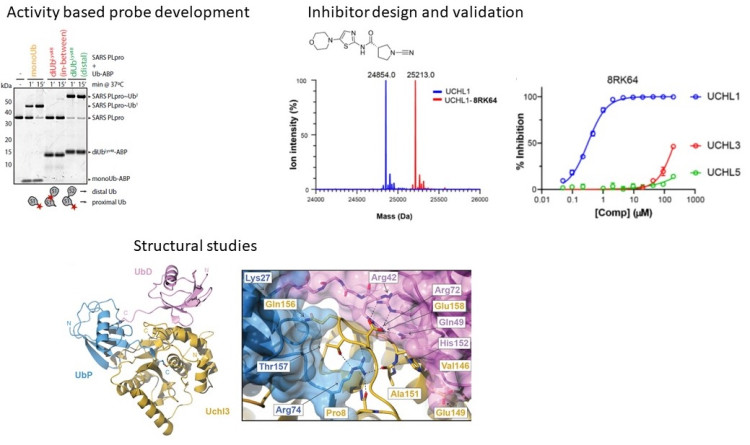
Based on the total chemical synthesis of ubiquitin, we developed a fluorescence polarization assay to assess ubiquitinated peptide selectivity of DUBs which turned out to be crucial in the elucidation of Ub linkage specificities of OTU DUBs. This technique was further extended to Ub-like proteins to assess bacterial effector proteases and the ISG15-specific protease USP18. This work laid the basis for the development of full-length diUb FRET probes with which the Ub linkage specificity of DUBs can be quantified. Such different assaying techniques are now further used to develop small-molecule cell permeable DUB inhibitors and activity-based probes.
The Ovaa lab

The Mulder, Geurink & van der Heden labs are the continuation of the group founded by our dear colleague and friend professor Dr. Huib Ovaa who passed away from prostate cancer on the 19th of May 2020. The summer before Huib became ill, and in first instance it appeared that he had attracted a relatively harmless infectious disease. Unfortunately, it became soon apparent that he suffered from a far more severe disease. In that year, Huib has undergone a variety of treatments, and has done so with his usual optimism. He continued to work as much as possible and tried to keep the burden of his fight against cancer away from the people around him as much as possible. Huib was 46 years of age.
His group studied enzymatic action in the ubiquitin proteasome system (UPS) and MHC class I antigen presentation using a combination of techniques including chemical synthesis and biochemistry. We are specialized in the design of assays and dedicated reagents that allow the study of protein ubiquitination and deubiquitination and that facilitate drug discovery efforts. We have an interest in the validation of potential therapeutic targets within the UPS with a focus on oncology while some are of relevance for the treatment of neurodegeneration. Approaches that the lab take include high-throughput screening using our acoustic dispensing system, hit validation and medicinal chemistry. Specific topics include modulation of the 26S proteasome activity (both activation and inhibition) in cells by small molecules as well as the development of dedicated PROTAC reagents. The lab is fully equipped for (high throughput) peptide synthesis, cell biology, biochemistry, immunology and high throughput small molecules screening, but also for medicinal chemistry. Next to screening and medicinal chemistry the lab has a longstanding interest in the development of probes that enable studies of the ubiquitin system. For example the lab has developed methods for the synthesis of ubiquitin and various ubiquitin-like reagents and probes.
Curriculum Vitae:
Huib carried out his PhD in the laboratory of prof. Jacques van Boom and spent part of his time in the lab of prof. Blechert at the TU Berlin. He was trained in the use of organometallic reactions on carbohydrate derived synthons to constract carbasugars. After receiving his PhD he moved to the lab of prof. Hidde Ploegh at Harvard Medical School to get familiar with biochemistry and immunology. After two years as postdoc and then some time as instructor in pathology he moved to the Netherlands Cancer Institute in 2004 to start a lab with a focus on chemical biology, specializing in the chemistry of ubiquitination, proteasomal proteolysis and antigen presentation. Since 2016 the lab is at Leiden Unverstity Medical Centre and part of the newly created department of Cell and Chemical Biology, where the lab has a focus on basic discovery, probe and assay development, and high throughput screening and medicinal chemistry.
Huibs career was as follows:
2016 – 2020 professor Leiden University Medical Centre
2016 – 2020 professor Leiden University
2012 – 2015 extraordinary professor Leiden University
2010 – 2013 0.1FTE C.S.O. UbiQ Bio B.V.
2009 – 2016 Tenured staff member NKI
2005 – 2012 Assistant Professor, Leiden University, the Netherlands
2004 – 2009 Junior group leader, NKI
2004 – 2005 Guest Researcher, Leiden University
2003 – 2004 Instructor in Pathology, Harvard Medical School, Boston, USA
2001 – 2003 Post-doctoral fellow, Harvard Medical School, Boston, USA
.jpg)