Background
This part of our research focusses on type 2 diabetes. Type 2 diabetes is responsible for about 90% of all diabetes cases and is caused by a combination of insulin resistance and insufficient insulin secretion. In addition to genetic susceptibility, overweight and an inactive life style are major risk factors. Our studies use a combination of modern omics techniques, data science and bioinformatics to identify biomarkers for disease progression.
Patients with type 2 diabetes follow different trajectories of glycaemic progression once the disease has developed. Whilst the majority show no or limited glycaemic progression, about 10-30 percent are unable to achieve HbA1c levels below the glycaemic target despite intensive treatment. Whereas especially this group has an increased risk of diabetic complications. Early identification of the persons at risk may help to redirect our clinical resources to those who may benefit the most thereby facilitating value-based healthcare. At present there are however no good instruments to identify those at risk of progression at an early stage.
As indicated above clinical studies have not yet yielded strong predictors of glycaemic progression or development of diabetes complications.
Multiomics and big data science
In our group we are investigating the role and utility as biomarkers of various omic datatypes in diabetes, diabetes progression and complications. We apply a combination of multi-omics measurements, data science and functional studies. This research is performed in close collaboration with Professors JW Beulens and PM Elders from Amsterdam UMC, Location VUmc. We use data from the Hoorn studies and the Hoorn Diabetes Care System Cohort (DCS) for our studies. These cohort studies contain longitudinally collected clinical data and biomaterials of thousands of healthy and affected persons (www.hoornstudies.com).
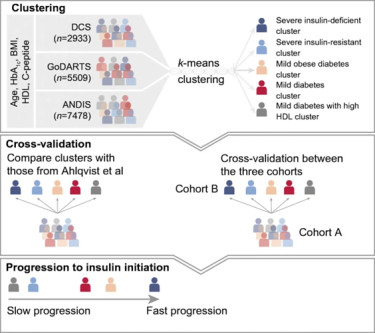
During the past years we have shown that various metabolites associate with diabetes, diabetes progression and complications such as Diabetic Kidney Disease (DKD) (BBMRI-NL). In addition, we participate in research to identify novel genetic predictors of drug response for commonly used diabetes drugs (Metformin, SU and DPP4-inhibitors). In two EU-funded projects (IMI-DIRECT and IMI-RHAPSODY) we investigated the utility of various omics measurements (Genomics, lipidomics, metabolomics and peptidomics) in risk stratification for type 2 diabetes development and progression. In these and other studies, we showed that five clusters of type 2 diabetes each have their own distinct molecular signatures (PMID: 34376475, 34110439, 36826505). Thereby, providing a better understanding of the heterogeneity of type 2 diabetes.
Furthermore, we identified biomarkers belonging to three different chemical classes that associate with rapid glycaemic progression, defined as the time to insulin use (PMID: 37137910). Using machine learning techniques we also showed that a set of biomarkers modestly improved prediction of rapid glycaemic progression on top of clinical biomarkers (PMID: 38374450).
Picture from: https://link.springer.com/article/10.1007/s00125-021-05490-8
Non coding RNAs
More recently, we also started projects investigating the role of circulating small and long non-coding RNAs in diabetes. In these studies we have identified a number of small and long non-coding RNAs that are associated with diabetes subtypes and or diabetes related complications. MicroRNAs are the most well-known members of the small non-coding RNAs and numerous studies have demonstrated their involvement in the (dys)regulation of gene expression in diabetes. However, there are many more small non-coding RNA biotypes and their function in diabetes is often unknown. We recently showed that, beyond several miRNAs, other small ncRNA biotypes such as snoRNAs, vault RNAs and piRNAs are associated with diabetes subtypes and/or diabetic kidney disease. For instance, the piRNA-019825 is associated with diabetic kidney disease in the Hoorn DCS cohort and we also showed that plasma levels of this piRNA show a strong correlation with the plasma levels of proteins that are known to predict future diabetic kidney disease risk. Using a combination of data science and functional studies we are currently further exploring the function of several of these small ncRNAs. In addition, we study their potential and role in disease treatment.